For this new edition of BioHackathon Europe, organised by ELIXIR Europe, participants and the ELIXIR France delegation travelled to Berlin from 3 to 7 November 2025. This international collaborative event brought together around 175 (in person) and 290 (remotely) researchers, bioinformaticians and developers to work on projects aimed at addressing research challenges in bioinformatics. This annual hackathon is a place where projects are accelerated to innovate in areas such as data integration, data science and the application of FAIR (Findable, Accessible, Interoperable, Reusable) principles, where participants implement their ideas during intensive coding sessions.
This Biohackathon has enabled and continues to enable projects to generate new developments (integration of prototypes, proof-of-concept proposals or production drafts), test new versions, gather information for new features, reach new users, and receive expert advice in various fields.
This year, our teams submitted four projects among the 31 selected for the BioHackathon, once again demonstrating the strong involvement of the French node teams.
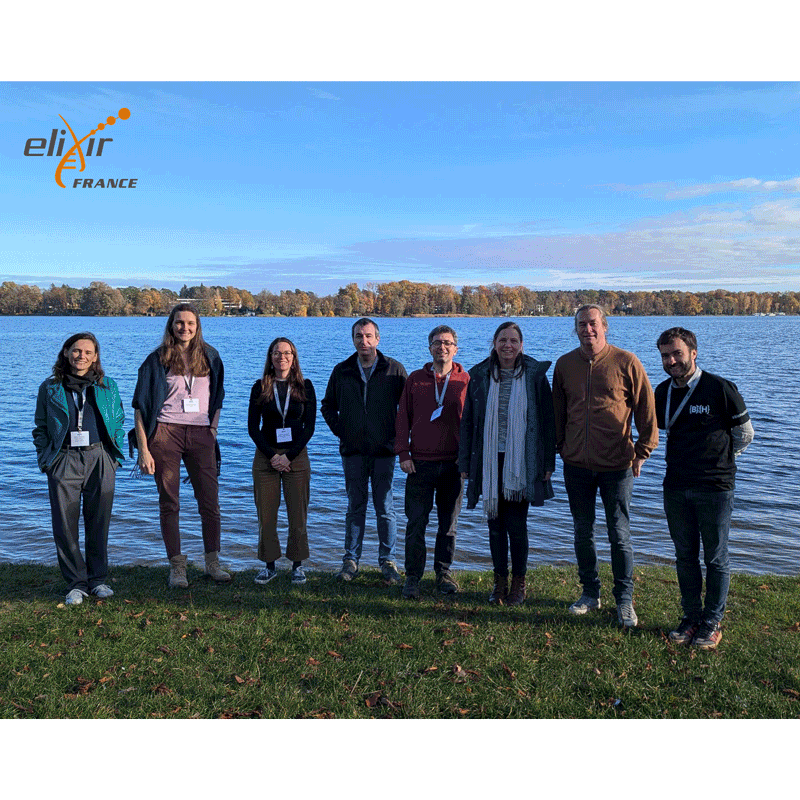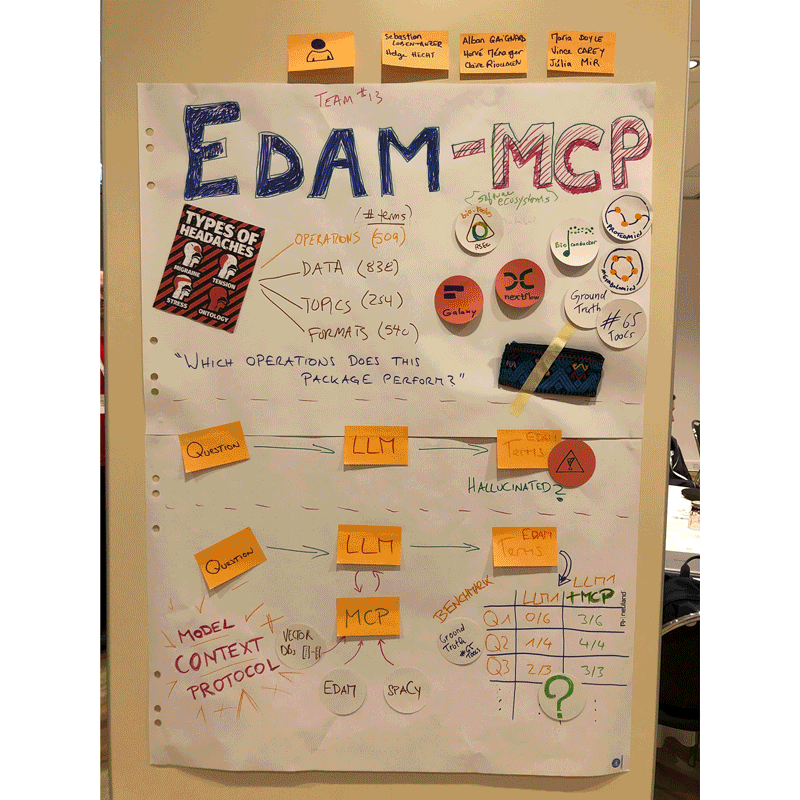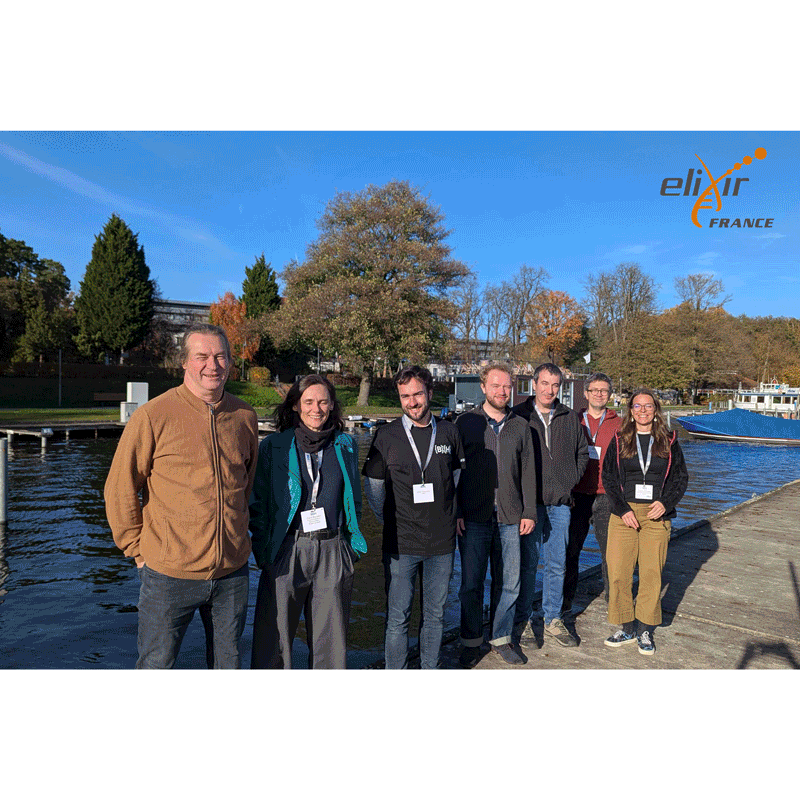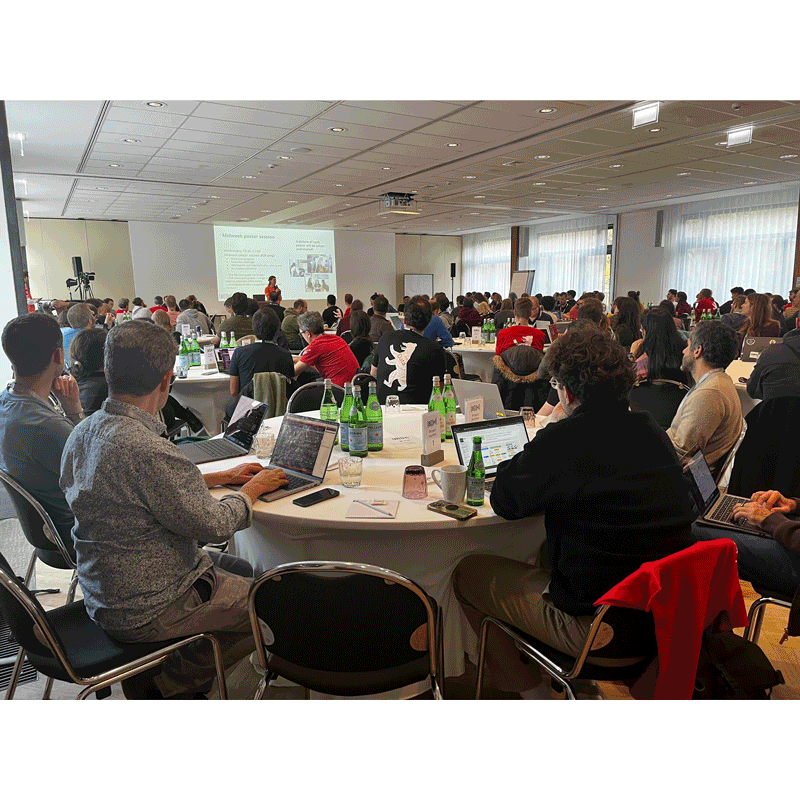
Project 13 – Improving package annotation in metabolomics and proteomics via robust, ontology-driven LLM integration by Claire Rioualen, co-led with Sebastian Lobentanzer (ELIXIR-DE) and Helge Hecht (ELIXIR-CZ)
Our project focused on developing EDAM MCP (Model Context Protocol), a tool to facilitate the annotation of bioinformatics resources with the EDAM ontology. In order to test the validity of the prototype, we gathered a set of reference tools and annotations in the field of metabolomics, showing convincing results. This BioHackathon enabled us to bring together collaborators with varied expertise from across Europe and the United States.
Project 14 – Metabolomics and Proteomics file format interoperability fest by Franck Giacomoni
This project brought together around forty participants around the mzTab-M format during the ELIXIR Europe 2025 BioHackathon in Bad Saarow and at a satellite event in Clermont-Ferrand. The team first mapped the mzTab-M software implementations, collected real metabolomics and lipidomics datasets, and automated their validation via the jmzTab-M validator and GitHub workflows in the HUPO-PSI repository. A ‘cross-interoperability’ campaign then tested around twenty combinations between producer tools (mzmine, MS-Dial, xcms, LipidDataAnalyzer, Progenesis QI, MetaboScape, MASSter) and consumer platforms (GNPS, LipidCompass, MetFamily, MetaboLights, MetaboAnalyst), with a majority of successful conversions. This work enabled bugs to be fixed, structured tickets to be opened, and discussions to be held on consolidating common libraries in Python and R, facilitating their adoption and long-term maintenance. The project also strengthened the integration of mzTab-M into ecosystems important to the community, such as Galaxy, PeakForest, EDAM, and bio.tools. Finally, a dedicated group outlined the necessary changes to the specification in order to, for example, better manage multifactorial experimental designs or ambiguities in metabolite annotation.
‘This BioHackathon has enabled the community to progress by both strengthening the various implementations of the mzTab-M format and fostering informed discussions on the necessary future developments of this format’s specifications, laying the groundwork for more reproducible, comparable and interoperable metabolomics and lipidomics.’
Project 16 – MiCoReCa (Microbiome Community Resource Catalogue): Towards centralised curation and integration by Bérénice Batut, co-led with Nikolaos Strepis (ELIXIR-NL) and Vivek Ashokan (ELIXIR-FR)
Notre projet a développé un processus de collecte, filtrage et curation des ressources ELIXIR dédiées au microbiome (outils, workflows, packages bioconda). Nous avons défini des mots-clés ciblés, extrait et initié la curation manuelle, et identifié des termes EDAM manquants. Nous collaborons maintenant avec la communauté ELIXIR Microbiome pour valider ces mots-clés, curer les ressources et proposer de nouveaux termes EDAM. Ce cadre méthodologique, conçu pour être réplicable, sera étendu aux communautés biodiversité et single-cell omics. Ce travail s’aligne avec les projets MUDIS4LS et Cloud4SAMS, ainsi qu’avec le Groupe de Travail « Catalogue des ressources » de la communauté Microbiologie (C4) de l’IFB.
Project 21 – Opening the TMD: making it more Usable, Visible, and Connected by Olivier Sand, co-hosted with Nina Norgren (ELIXIR-SE) and Eleni Adamidi (ELIXIR-GR)
The aim of our project was to develop an API and widgets for the Training Metrics Database, which collects demographic data and information on the quality and impact of training courses offered by ELIXIR nodes. The API works by retrieving aggregated data from all nodes. The next step is to add the functionality to add and retrieve data from a specific node. Configurable code snippets can be integrated into websites to view aggregated data across all nodes. The functionality to do this for data from a single node is currently under development. The BioHackathon also helped to improve the user interface.
