Total computing capabilities and performance
Total compute
27896
#CPU HT*
Total storage
15
#PB
Total RAM
45
#PB
Total GPU
44
#Card
* CPU HT = hyperthreaded
Choosing your computing solution
All of the IFB’s computing and storage offerings are supported by the NNCR (National Network of Computing Resources).
- Platforms participating in the clusters : ABiMS , BiRD , GenoToul , GenOuest , IFB-BiGEst-Cluster , IFB Core , MicroScope, Migale , South Green
- Platforms participating in the Cloud federation : AuBi, Bilille, BiRD, GenOuest, IFB-BigEst-Cloud, IFB-core, Prabi, CBP-PSMN
These features and capabilities allow us to meet specific uses and analysis needs .
Features and uses
Cluster
- HPC (High Performance Computer) type infrastructure
- Multiple access interfaces: SSH, Galaxy and other web portals
- Bioinformatics resources: general and specialized
- Software environments via Conda and Singularity already configured
- Solution adapted to biologist and bioinformatician users
- User Expertise Levels: From Novice to Expert
Cloud
- On-demand provision of computing resources and reference data
- Possibility of deploying appliances (preconfigured machines) or installing your own bioinformatics infrastructure.
- High flexibility and total control over the environment
- On-demand resources
- User Expertise Levels: From Novice to Expert
Types of analysis
Cluster
- Intensive computing requiring many computing cores simultaneously
- Standardized analyses with already installed tools
- Work requiring access to large volumes of shared storage
Cloud
- Specific bioinformatics analyses requiring preconfigured software environments
- Projects requiring custom configurations
- Testing new tools or developments
Technical capabilities
Cluster
- 4300 cores (hyperthreaded)
- 2 PB of storage
Cloud
- 6,000 computing cores
- 28 terabytes (TB) of memory
- Resources distributed between 7 sites
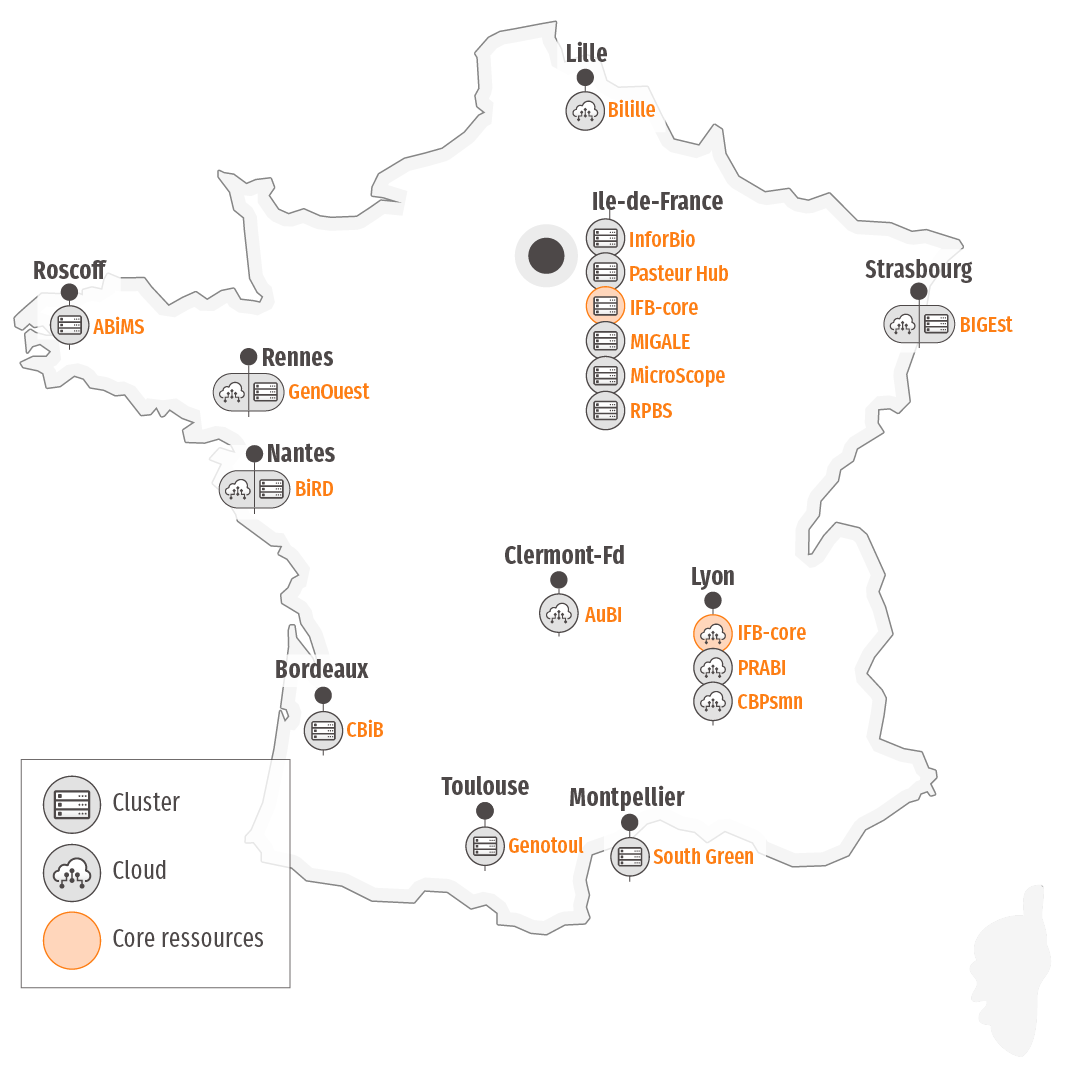
Geographic location
IFB’s computing and storage offering is deployed through our network of member platforms.

All of the IFB’s computing and storage offerings are supported by the NNCR (National Network of Computing Resources).
The National Network of Computing Resources - NNCR
Community services are provided by the NNCR, which includes all the hardware (IFB-core + regional platforms) that contributes to ensuring the service offering. The NNCR relies on a distributed infrastructure consisting of 8 clouds and 6 clusters spread across 13 regional centers, and 2 centralized resources: the IFB-core servers (cloud in Lyon, cluster in Orsay), and nine high-performance computing platforms located in different regions. This network is open to French and international research communities, both public and private, in the field of life sciences.
The IFB also plays a key role in the national strategy for pooling and modernizing scientific digital infrastructure. Finally, at the ELIXIR level, this service contributes to the maintenance of Biocontainers and the coordination of the Galaxy community within ELIXIR.
Compared to data processing in other scientific fields, the specific characteristics of biological data make them dependent on a very specific hardware architecture, software environment, and data collections that are currently not available in national computing centers. To address these challenges, the IFB has organized its services by decentralizing physical, logistical, and human resources.
Such decentralization of resources offers at least four main advantages:
> The IFB Cloud
The cloud federation offers a set of preconfigured environments, allowing scientists and bioinformaticians to choose the configuration best suited to their analyses. These virtual bioinformatics environments developed by certain members of the IFB community are registered in the Biosphère catalog. Scientists and engineers can launch their own virtual environments with dedicated resources that can be adapted to their needs without interfering with other users. This activity is constantly growing, and the IFB’s French cloud has already been used for numerous scientific analyses and training sessions, workshops, hackathons, and several recurring sessions at scientific schools or universities.
An account offers variable space (from 25 GB to 3 TB per virtual machine for the duration of job execution but deleted when the virtual machine is shut down) to which will soon be associated a persistent storage space shared between the virtual machines of a user or group.
You can also use the IFB cloud as a support for your training. To find out more
> Clusters in the region
All clusters on our regional platforms are dedicated to regional and thematic users.
> The IFB-core cluster
What is the IFB-core cluster?
The Core Cluster is IFB’s national computing infrastructure. It aims to meet the computing needs of all communities in the health and biology fields, with a focus on users who do not have local computing resources.
The infrastructure is open to all users with an academic email address in France or in one of the ELIXIR member countries .
The Core Cluster is accessible via three modalities:
The Core Cluster is administered collaboratively. More than six engineers from five IFB platforms build and contribute to the project daily. To manage the multiple contributions, they are overseen by CI mechanisms (Ansible + Gitlab runner) connected to a common code repository (Gitlab).
Complete Cluster documentation: https://ifb-elixirfr.gitlab.io/cluster/doc/
Terms of Use: https://ifb-elixirfr.gitlab.io/cluster/doc/terms-of-usage/
Services
Accounts management
All academic users could request an account to access Core Cluster via our account registration and management portal : https://my.cluster.france-bioinformatique.fr. Beware, this account is usable only for the Core Cluster and won’t be recognize by other IFB services.
Submission of computer tasks
The primary use of the cluster is with SLURM from an SSH console.
If you are new to SLURM, please read the Core Cluster documentation and tutorial to learn how to submit your first jobs:
https://ifb-elixirfr.gitlab.io/cluster/doc/slurm/slurm_user_guide/
Interactive web portal
The Open Ondemand web portal allows you to run interactive tools like RStudio or Jupyterlab on cluster resources via a simple web interface. Find the overview page , documentation and dedicated video .
Galaxy
The IFB Core Cluster provides the computing resources for the French instance of Galaxy: usegalaxy.fr
usegalaxy.fr offer a large panel of bioinformatics software usabled in line. Some software are also usabled from thematics sub-domaines such as metabolomic, singleCell, covid19, etc.
Technical support
The support.cluster.france-bioinformatique.fr portal allows cluster users to contact our support team for any technical requests, including:
* Support for usegalaxy.fr is available on the IFB community forum
Support for the bioinformatics community
A community forum allows you to exchange ideas with biologists and bioinformaticians on bioinformatics: uses and options for a tool, setting up a workflow around a specific theme, etc.
Training hosting
The IFB Core Cluster can provide computing resources for your training session. Our support team offers a variety of options, including creating temporary generic accounts and reserving cluster resources.
To request cluster resources for your training, complete the request form .
Contribute to the IFB-core Cluster TaskForce
If you have skills in bioinformatics tools or system administration, join the IFB Core Cluster TaskForce to deploy your tools or contribute to the management of the IFB infrastructure and become part of the legend. Please do not hesitate to contact us by sending an email to:
contact-nncr-cluster@groupes.france-bioinformatique.fr Otherwise, if you wish to be trained in the technologies used on the infrastructure, training/tutorial sessions are offered regularly.
Continuous Integration (CI) & Collaborative Work
Resource administration is carried out collaboratively. In order to manage multiple contributions, these are managed by a continuous integration mechanism connected to a common code repository.
Traceability & Secure Contributions
All installation, configuration and maintenance actions must be traceable as much as possible. This is in order to:
We chose to use a Git repository hosted on GitLab. Git meets all our traceability needs. The GitLab interface provides us with a space to exchange information and offers the ability to work with Merge Requests (Pull Requests) and host our own CI job runners. These MRs require the inclusion of a code review phase before going live. Each change or addition is validated by a peer review to avoid errors and ensure that at least two people are aware of it.
Task Force Members: