Include the following sentence in the “Funding” or “Acknowledgement” section of publications:
Project description
Analysing the impact of the human microbiome on health is a scientific and medical challenge that raises both technological (massive, scattered and heterogeneous data) and ethical (potentially identifying nature of microbiome data) issues. In this context, the Cloud4SAMS targeted project of the France 2030 research programme ‘Food Systems, Microbiomes and Health’ aims to deploy a digital computing and storage infrastructure enabling scientists to exploit microbiome and health data in a secure IT environment. The architecture and implementation are based on the expertise and standards developed by IFB (bioinformatics resource catalogue, FAIRisation and data brokering tools) and by the ELIXIR network (bio.tools catalogue, workflow hub, EDAM ontology, Federated European Genome Archive).
The datasets produced by microbiome projects (particularly those funded by the France 2030 SAMS research programme) and the software tools and workflows for processing these data will be indexed in the Cloud4SAMS catalogue (WP2) catalogue and will serve as building blocks for defining deployment recipes describing all the procedures for instantiating a virtual machine in a secure or academic cloud (WP3). Access to this data is facilitated by an interface that manages requests to each project’s access committees, authorisation validation, ad hoc data extraction and transfer to secure spaces (WP4). This access will comply with current regulations on the use of health data (including microbiome data). WP6 of the Cloud4SAMS project will thus provide recommendations, procedures and guidelines for sharing sensitive data under ad hoc conditions. The project also includes an important component dedicated to dissemination and training (WP5): support for data producers in assessing data quality, improving metadata, and submitting to reference repositories; training users in IT resources (catalogue, access portal, secure environments) as well as in ethical and legal issues relating to personal data.
Taking into account financial resources and deadlines, Cloud4SAMS will limit itself to implementing a proof of concept (POC) aimed at deploying all the basic building blocks and testing the system in a few selected use cases (WP7) based on data produced by partner teams or available in the public domain.
WP1 – Project coordination and management
Co-responsables : Nicolas Pons (INRAE), Claudine Médigue (CNRS)
Goals :
- Preparation of contractual documents.
- Implementation and management of the governance framework.
- Organisation of consortium meetings, financial and scientific reports.
- Establishment of cross-functional working groups for national microbial bioinformatics projects led by the IFB.
- Definition of actions for the sustainability of the Cloud4SAMS platform beyond its initial funding.
WP2 – Digital resources catalogue
Co-responsables : Nicolas Pons (INRAE), Claudine Médigue (CNRS), Samuel Chaffron (CNRS)
Goals :
- Provide a federated catalogue of computational resources as a standardised generic toolkit for microbiome data management and basic processing:
- data produced by microbiome projects;
- software, tools and workflows;
- IT facilities suitable for processing microbiome and health data.
- Enable the definition of deployment recipes describing the procedures for instantiating a virtual environment in a secure cloud (WP3).
- Promote compliance with national and international standards by leveraging existing resources (e.g. the IFB catalogue of bioinformatics resources, the ELIXIR bio.tools and WorkflowHub repositories).
WP3 – Secure digital environment
Co-responsables : Christophe Blanchet (CNRS), Antoine Fraboulet (INRIA)
Goals :
- Design, deploy, and test a highly secure cloud environment for data processing for researchers working with microbiome and health data, in compliance with regulations governing sensitive data.
- Participate in the implementation of the necessary security technologies to guarantee or improve the level of confidentiality, integrity and availability of data processed in the targeted NNCR IT facilities of the IFB.
- Improve and enrich existing environments (IFB Biosphere catalogue) in order to meet the specific requirements of the Cloud4SAMS project.
- Evaluate the chain implemented, from deployment requirements (virtual machines, targeted secure infrastructure, data transfer, etc.) to researchers’ ability to securely access their environments and process their data through the use cases defined in Cloud4SAMS.
WP4 – Data access management
Co-responsables : Alban Gaignard (CNRS), David Salgado (Inserm)
Goals :
- Enable simplified access to microbiome data through a user-friendly interface that manages:
- applications to the access committees for each project;
- the approval or rejection of authorisations;
- ad hoc data extraction;
- and their transfer to secure areas.
- Develop an API ensuring interoperability with federated resources.
- Ensure the traceability of all transactions.
- Identify the best data transfer procedures.
WP5 – Training and dissemination
Co-responsables : Hélène Chiapello (INRAE)
Goals :
- Development of training materials and organisation of training sessions on the various components of Cloud4SAMS Organisation of hackathons to involve the community in the adoption and improvement of Cloud4SAMS.
- Organisation of data brokering in permanent, secure data warehouses that comply with data regulations.
- Organisation of scientific communication (publications, conferences, webinars, workshops) to promote interest in open science and data sharing in a secure, trusted ecosystem that will accelerate research in the field of the microbiome and health.
WP6 – Ethical, legal and social considerations
Responsable : Nathalie Gandon (INRAE)
Goals :
- Documenting the regulatory framework:
- Make this framework accessible;
- Provide standard documents (protocols, information sheets, consent forms, etc.);
- Propose an INRAE organisation in coordination with the relevant departments (DAJ, DSB, DPO, etc.);
- Train project leaders.
- Supporting project leaders funded by the France 2030 SAMS research programme on regulatory compliance related to research involving human subjects.
- Qualifying microbiota data:
- Analyse whether they can become directly identifiable or not;
- Explain the security measures required for their processing.
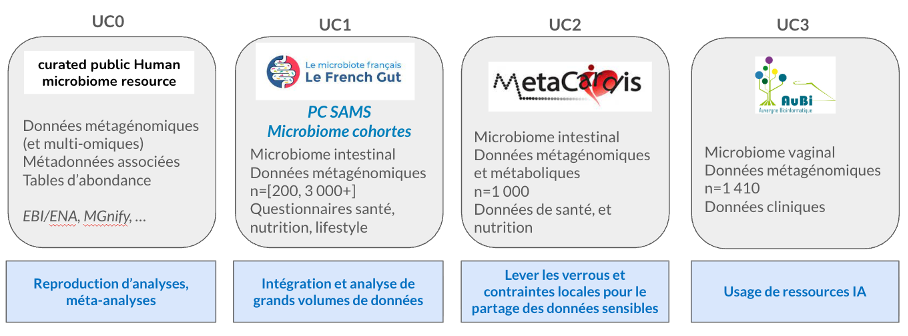
In order to ensure that developments to the Cloud4SAMS platform are perfectly suited to the needs of future users, four use cases have been identified (initially). These should enable:
- Test the articulation and performance of the infrastructure bricks;
- Describe and implement standard deployment recipes in appropriate processing environments.
- Remove barriers to data sharing and ensure trust-based sharing;
- Identify needs and integrate new reference resources related to WP2 (databases, tools, methodologies, etc.).
These use cases are described in the figure below:
Details of the various use cases will be available very soon. Please check back on this page at a later date.
26 September 2024 – Project launch
6 February 2025 – 1st PEPR SAMS Day
The Cloud4SAMS project was presented at the first annual scientific day of the France 2030 Food Systems, Microbiomes and Health research programme (PEPR SAMS) held on 6 February 2025.
2028 – End of the project
All publications affiliated with the Cloud4SAMS project can be found in HAL.
19 and 20 November 2025 – Development team hackathon
The Cloud4SAMS development team met on 19 and 20 November in Paris for a hackathon dedicated to innovation and the evolution of the project. Much more than just an intensive work session, this hackathon provided an opportunity to test new ideas, deploy new strategic developments necessary for the Cloud4SAMS project, and strengthen ties within the team of developers.
13 November 2025 – Presentation of the project during the annual PEPR SAMS day
16 October 2025 – Annual project meeting
The annual meeting of the Cloud4SAMS project, attended by three representatives from the ANR, four representatives from the PEPR SAMS management team, members of the Copil and the project co-coordinators, took place via videoconference on 16 October 2025. Claudine Médigue and Nicolas Pons presented the progress made and the revised roadmap, which were very well received by the ANR and PEPR SAMS: they were unanimous in their praise of the quality and clarity of the presentations and the management of the Cloud4SAMS project stages.
One of the main points of the meeting concerned regulatory aspects and data protection (WP6), which raise many questions that go beyond the scope of Cloud4SAMS. The PEPR management would like to receive a summary note on the issues and difficulties raised by WP6 in order to share and communicate this information at a multi-institutional level. In addition, the Cloud4SAMS co-coordinators are encouraged to organise a workshop on data management and protection for PEPR SAMS projects, as well as other human microbiota data analysis projects.
6 and 7 May 2025 – Project workshop (news available on the PEPR SAMS website)
The Cloud4SAMS targeted project consortium met on Tuesday 6 and Wednesday 7 May 2025 for a workshop organised at Inserm headquarters in Paris. The aim of this workshop was to identify and prioritise developments for the Cloud4SAMS secure platform through a detailed presentation of the requirements for various identified use cases.
The action plan drawn up following this workshop includes expanding the IFB Cloud’s catalogue of metagenomic analysis methods and organising tutorials and workshops on how to use the Biosphere environment for consortium members. At the same time, the deployment of Biosphere appliances will be tested in Arcana (Inria HDS Cloud). Initiatives are also being launched to pool metagenomic workflows, improve data discoverability, and develop two prototypes based on concrete use cases.